Project Details
African populations are characterized by a high level of genetic diversity owing to a large number of variable genes and alleles (Jorde et al., 2000; Tishkoff & Williams, 2002; Cavalli-Sforza et al., 1997; Jakobson et al., 2008). Patterns of genetic variation in the modern African population are influenced by a demographic history that includes changes in population size, admixture and locus-specific forces such as natural selection, recombination and mutation. To date, few genetic studies of structural variation of genes across ethnically diverse populations have been conducted (Conrad et al., 2007). There are currently no studies within and between ethnically diverse African populations.
South Africa carries a higher burden of HIV infection than any other country in the world and embodies a rich collection of ethnic backgrounds. The major ethnic groups include the Bapedi, Basotho, Ndebele, Swati, Tsonga, Tswana, Xhosa, Venda and Zulu. The genetic substructure of these populations was assessed by studying the Y-chromosome and autosomal DNA of these populations (Lane et al., 2002; Matchell, 2010). The Y-chromosome data demonstrated that people speaking these ethnic languages cluster into three specific groups (Tswana/Sotho, Nguni and Venda). Thus people belonging to these linguistic groups also show genetic differences. Because of their genetic variability, these populations are also likely to carry variable host cell genes (restriction and other factors) that may play a crucial role in viral replication.
As described above, host cell genes such as the C-C chemokine receptor type 5 (CCR5) play a major role in HIV-1 infection and disease progression. One study described how polymorphisms within the open reading frame as well as the regulatory regions could influence the amount of CCR5 expressed on the cell surface, and hence individual susceptibility to HIV-1 infection (Picton et al., 2010). There are currently no studies describing CCR5 expression levels in ethnically diverse populations of South Africa and how expression levels may be influenced by variability. In addition, mutations in the V3 loop, a major determinant for tropism, could affect syncytium formation, viral infectivity, neutralization, replication efficiency and host cell tropism.
A fundamental concept in the biology of viral restriction factors is that HIV-1 generally evades their potent inhibitory activities in human cells, thereby allowing viral replication to proceed efficiently (Malim & Bieniasz, 2012). Many HIV restriction factors and their counteractants have been described. Of importance in this study is the vif protein, which antagonizes apoliprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins, particularly APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H (Sheehy et al., 2002).
These restriction factors exhibit different mechanisms to block viral replication. Since restriction and antagonizing factors interact with each other through direct protein-protein binding, both restriction factors and their viral counterpart proteins often show the signature of relatively rapid evolution. This evolution drives selection for beneficial mutations in restriction factor genes and their viral antagonists. As a consequence of positive selection, even phylogenetically similar species are likely to differ in terms of restriction factor functionality. Many APOBEC3 genes, including APOBEC3G, H and D have evolved under positive selection for millions of years in primates (Dugga et al., 2011; OhAinle et al., 2008; Sawyer et al., 2004). Within humans, several APOBEC3s are known to possess common polymorphisms that render them defective, reduce antiviral activity, and increase sensitivity to HIV-1 vif. Furthermore, such polymorphisms are common in some populations, while not present in others.
Population-based studies have tried to establish a relationship between host factors such as APOBEC3 as well as CCR5 polymorphism, expression or activity and the rate of disease progression (Pace et al., 2006; Vetter et al., 2009; Vazquez et al., 2009). Most studies have focused on Caucasian populations. In contrast, little information is available for the effects of variation in these genes in African populations, where the HIV epidemic has expanded at an alarming rate. Although several population studies have focused on African Americans, these do not give us a complete picture of the potential variation in Africans, though the studies can be a good guide from which to base additional studies. A more comprehensive analysis involving different African ethnic groups will likely provide a better understanding of the mechanisms underlying host pathogen interactions, especially in view of the fact that African Americans are primarily infected with HIV subtype B, which is rarely seen in Africa.
The main objective of this study is to characterize the variability of CCR5 and APOBEC3 genes and to determine how they influence HIV-1 evolution and genetic diversity in ethnically diverse populations in Limpopo province of South Africa. This study will be correlated with sequence analysis of the HIV-1 V3 and vif regions as well as analysis of HIV “editing”.
Project History
Initially the idea of the project was to look at restriction factors, in particular the APOBEC3 genes and their countreactant (HIV-1 vif) and how polymorphisms within these genes affect their interactions with the application of the latest sequencing technology (Next Generation Sequencing). We are currently also looking into the CCR5/CXCR4 and V3 loop host-viral interaction. We have of recent characterized polymorphism in APOBEC3D, APOBEC3F, APOBECG, APOBECH, CCR5, HIV-1 vif and V3 loop gene and new hypothesis are being derived, such as how these polymorphisms affect replication and contribute to the overall HIV-1 diversity.
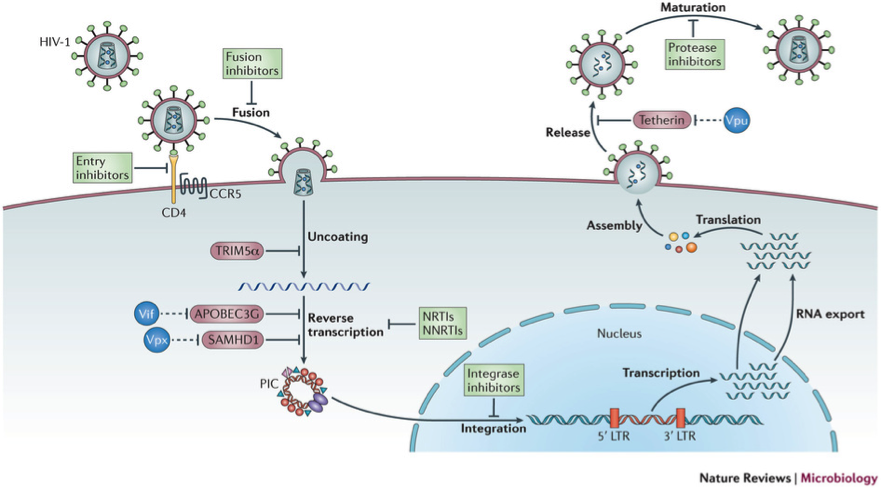
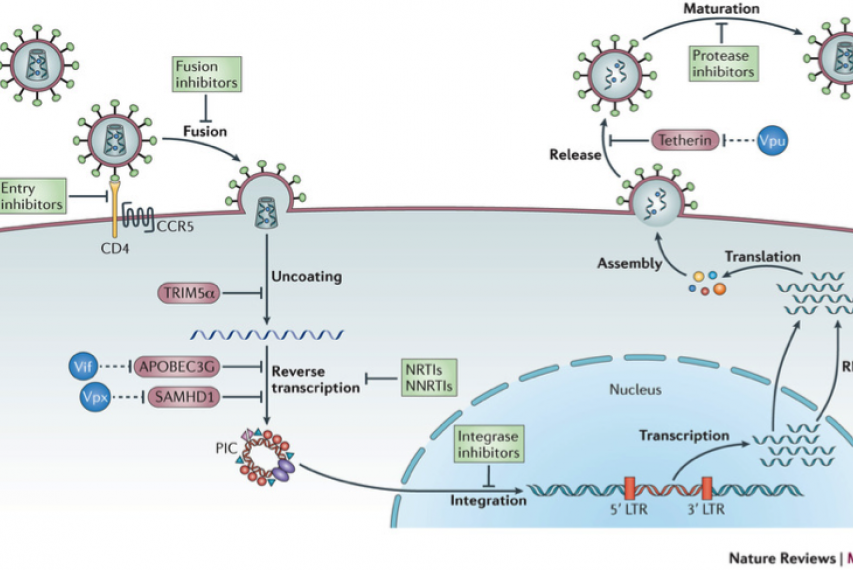
Figure 1: The figure illustrates the main steps in the HIV-1 replication cycle. The major families of antiretroviral drugs (green), and the step of the life cycle that they block, are indicated. Also shown are the key HIV restriction factors and their corresponding viral antagonists (vif, vpx and vpu in blue); CCR5, CC-chemokine receptor 5; LTR, long terminal repeat; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors. The major host –viral interaction of interest in the project are in red circles (Nature Reviews Microbiology 11, 2013).
Location
This work is taking place in the Limpopo Province of South Africa.
